Sequencing > Services
We offer a Full Service option for the standard applications listed below using validated library prep methods. We have an average turnaround time of about 4 weeks after the samples pass QC. If your sample doesn't reach the sample requirements of our standard protocols, we can suggest other protocols and we can continue with the experiment via our custom service mode or user lab mode.
You can find how to access our User Lab HERE
Please click on the application or protocol of your choice for a more detailed information (ie sample requirements, experimental design considerations).
Illumina Sequencing Pricing
How to submit samples for Illumina/Short Read Sequencing.pdf
How to submit Ready-made Libraries.pdf
Images are from https://www.illumina.com
Illumina Sequencing Pricing
How to submit samples for Illumina/Short Read Sequencing.pdf
How to submit Ready-made Libraries.pdf
STEP 1 : Project Registration.
Please register for a project in B-Fabric. All services in NGS require a project submission via B-Fabric, our project management system. Register for a project here.
As a project applicant/manager, you can add members to your project. These members can make orders and access project data. For more information about the data access rights, please click here.
STEP 2 : Assigning a Project Coach
You will be assigned a project coach, who will be your main FGCZ contact. Your project coach will invite you for a meeting to discuss your project in-depth together with one of our bioinformaticians.
This step is where you will settle on the best experimental design, library prep approach, sequencer and data analysis method for your research question. Your coach will review the project and forward it for approval.
STEP 3 : Create an order and sample registration
Once your project is approved, you may create an order and register the samples.
Instructions to fill out the form is here
How to submit samples for Illumina/Short Read Sequencing.pdf
How to submit Ready-made Libraries.pdf
STEP 4 : Offer Generation and Order Acceptance
Will will check the order and then make an offer. This step is where the FGCZ will commit to the deliverables and allow the you to check for the total cost of the experiment.
STEP 5 : Sample drop-off
If you agree to the terms in Step 4, you may bring your samples.
Do not bring or send samples which are not properly labeled with Bfabric IDs.
You can either bring the samples by hand or send them via post/courier.
We are glad to accept your samples between 9am to 5pm.
Our address: Functional Genomics Center Zurich, Sequencing Platform, Y59G90, Winterthurerstr. 190, 8057 Zurich SWITZERLAND
Please specify the name of your coach.
STEP 6 : Sample Tracking
You will receive an automated email (gfeeder) every time we reach a milestone in the sample preparation.
STEP 7: Data Delivery
Your data will be delivered via Bfabric.
STEP 8: Data Analysis
If you opted for bioinformatics analysis, our bioinformaticians will contact you about data analysis.
STEP 9: Invoicing
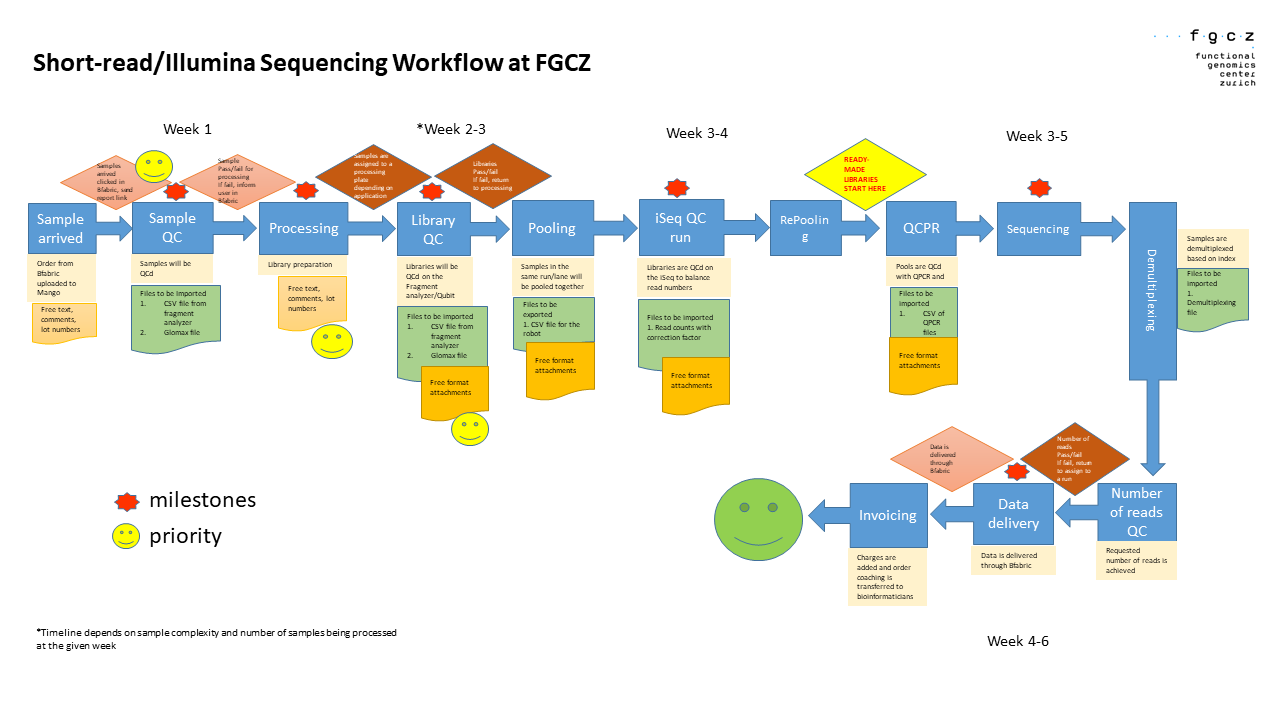
Our normal turnaround time is 4-6 weeks. Once we receive samples, we proceed directly with the processing of your samples. We receive double to triple the number of samples around June-August and November-December. This results in a slight increase in our turnaround time during these periods.
This illustration summarizes our workflow.
Project data can be accessed by active members of the project. Project managers can remove or give access rights in the B-Fabric project page.
Still have some questions? Please email sequencing@fgcz.ethz.ch
FGCZ apps
RNA Quality Explained
Library QC Explained
How to submit samples for Illumina/Short Read Sequencing.pdf
How to submit Ready-made Libraries.pdf
How to replace samples.pdf
Acknowledgements and Authorships
Introduction Slides
Back to FGCZ Main Page
NGS based Workflow Full Service
We offer a Full Service option for the standard applications listed below using validated library prep methods. We have an average turnaround time of about 4 weeks after the samples pass QC. If your sample doesn't reach the sample requirements of our standard protocols, we can suggest other protocols and we can continue with the experiment via our custom service mode or user lab mode.
You can find how to access our User Lab HERE
Standard Service Offerings, Applications and Sample Requirements:
| Applications | Protocols Offered | Optimum Input Requirements | Standard Sequencing Recommendations | |||||
| 1. mRNA Sequencing | Illumina TruSeq mRNA Stranded | 600 ng - 1 μg Total RNA, DNAse treated, column purified | 20M reads, single read 100 bp | |||||
| 2. Total RNA Sequencing | Illumina TruSeq TotalRNA Stranded (HMR) | 600 ng - 1 μg Total RNA, DNAse treated, column purified | 40M reads, single read 100 bp | |||||
| 3. Low Input RNA Sequencing | Clontech Picomammalian, SmartSeq2 | less than 10 ng Total RNA | 20-40M reads, single read 100 bp | |||||
| 4. Small RNA Sequencing | RealSeq | 1 ng - 1 μg Total RNA, DNAse treated | 10M reads, single read 75 bp | |||||
| 5. Single cell Multiomics Sequencing | 10X Genomics Single Cell, BD Rhapsody | 1’000 - 10’000 viable cells in a tube. | 50’000 reads per cell | |||||
| 6. Whole Genome Sequencing | TruSeq DNA Nano, TruSeq PCR Free, NEB Ultra II | 600 ng - 1'000 ng high MW genomic DNA | 30X coverage, paired end 150 bp | |||||
| 7. Low Input Whole Genome Sequencing | NEB Ultra II, Nextera XT | 1 - 10 ng high MW genomic DNA | 30X coverage, paired end 150 bp | |||||
| 8. Whole Small Genome Sequencing | Nextera XT | 1 - 10 ng genomic DNA | paired end 150 bp | |||||
| 9. Metagenomics | Amplicon Barcoding | inquire | inquire | |||||
| 10. Chip Seq | NEB Ultra | 10 ng Single ChIP enriched DNA or input DNA | 20M reads per sample, single read 100 bp | |||||
| 11. Ready-Made Libraries | 10 nM, pooled | custom | ||||||
| 12. Spatial Gene Expression | 10X Visium | FFPE | custom | |||||
Please click on the application or protocol of your choice for a more detailed information (ie sample requirements, experimental design considerations).
Illumina Sequencing Pricing
How to submit samples for Illumina/Short Read Sequencing.pdf
How to submit Ready-made Libraries.pdf
Short-read sequencers
| Sequencers | Ilumina NovaSeqX | Ilumina NovaSeq | Ilumina NextSeq 2000 | llumina NextSeq 500 | Ilumina MiSeq | Ilumina MiniSeq | Ilumina iSeq | |
| | 
| 
| 
| 
| 
| |||
| Flowcell | 25B, 10B, 1.5B | SP, S1,S2,S4 | P2, P3 | High Output, Mid output | High Output, Mid output | High Output, Mid output | ||
| Key Features | Patterned Flowcell | Patterned Flowcell | Patterned Flowcell | Non-Patterned Flowcell | Non-Patterned Flowcell | Non-Patterned Flowcell | Nanowells, CMOS | |
| Reads | 10B Reads per FC | S4 8 Billion | P3 1.1B | HO 350 Million | 25 Million | HO 22 Million | 4Million | |
| SP 650-800M | P2 400M | MO120 Million | MO 7 Million | |||||
| Max Read Length | 2X 150bp, 1X100bp | 2X150 bp, SP 2X250bp | 2X150 bp, 2X 250bp | 2X150 bp | 2X300 bp | 2X150 bp | 2X150 bp | |
| Approx Price per Gb | S4-12.10 chf, S1-25.9 chf | 46.5 chf | 196.8 chf | 282 chf | 925 chf | |||
| Key Applications | Large WGS, RNA Seq | Large WGS, RNA Seq | Single cell Sequencing | ATAC Seq, ChIP | Small WGS, Amplicons | Small WGS, QCs | Small WGS | |
| Single Cell, Spatial | Custom primers | small RNA | Targeted Sequencing, QCs | |||||
| Access Mode | Service | Service | Walk-in | Service, Walk-in | Service, Walk-in | Service, Walk-in | Service, Walk-in | |
| Location | Irchel Campus | Irchel Campus | GEML Honggerberg | Irchel Campus | Irchel Campus | Irchel Campus | Irchel Campus | |
| Irchel Campus | GEML Honggerberg | |||||||
Images are from https://www.illumina.com
Illumina Sequencing Pricing
How to submit samples for Illumina/Short Read Sequencing.pdf
How to submit Ready-made Libraries.pdf
Sample Submission Guidelines
STEP 1 : Project Registration.
Please register for a project in B-Fabric. All services in NGS require a project submission via B-Fabric, our project management system. Register for a project here.
As a project applicant/manager, you can add members to your project. These members can make orders and access project data. For more information about the data access rights, please click here.
STEP 2 : Assigning a Project Coach
You will be assigned a project coach, who will be your main FGCZ contact. Your project coach will invite you for a meeting to discuss your project in-depth together with one of our bioinformaticians.
This step is where you will settle on the best experimental design, library prep approach, sequencer and data analysis method for your research question. Your coach will review the project and forward it for approval.
STEP 3 : Create an order and sample registration
Once your project is approved, you may create an order and register the samples.
Instructions to fill out the form is here
How to submit samples for Illumina/Short Read Sequencing.pdf
How to submit Ready-made Libraries.pdf
STEP 4 : Offer Generation and Order Acceptance
Will will check the order and then make an offer. This step is where the FGCZ will commit to the deliverables and allow the you to check for the total cost of the experiment.
STEP 5 : Sample drop-off
If you agree to the terms in Step 4, you may bring your samples.
Do not bring or send samples which are not properly labeled with Bfabric IDs.
You can either bring the samples by hand or send them via post/courier.
We are glad to accept your samples between 9am to 5pm.
Our address: Functional Genomics Center Zurich, Sequencing Platform, Y59G90, Winterthurerstr. 190, 8057 Zurich SWITZERLAND
Please specify the name of your coach.
STEP 6 : Sample Tracking
You will receive an automated email (gfeeder) every time we reach a milestone in the sample preparation.
STEP 7: Data Delivery
Your data will be delivered via Bfabric.
STEP 8: Data Analysis
If you opted for bioinformatics analysis, our bioinformaticians will contact you about data analysis.
STEP 9: Invoicing
Full Service Workflow Process and Turnaround Times
Our normal turnaround time is 4-6 weeks. Once we receive samples, we proceed directly with the processing of your samples. We receive double to triple the number of samples around June-August and November-December. This results in a slight increase in our turnaround time during these periods.
This illustration summarizes our workflow.

Data Access Policy
Project data can be accessed by active members of the project. Project managers can remove or give access rights in the B-Fabric project page.
Still have some questions? Please email sequencing@fgcz.ethz.ch
Resources
Coverage CalculatorFGCZ apps
RNA Quality Explained
Library QC Explained
How to submit samples for Illumina/Short Read Sequencing.pdf
How to submit Ready-made Libraries.pdf
How to replace samples.pdf
Acknowledgements and Authorships
Introduction Slides
Back to FGCZ Main Page