Proteomics > Which service should I request? > Glycan / Glycoprotein analysis > Glycopeptide analysis
Questions?
Depending on the type of samples:
Some important points:
Glycopeptide analysis
Table of contents

General description
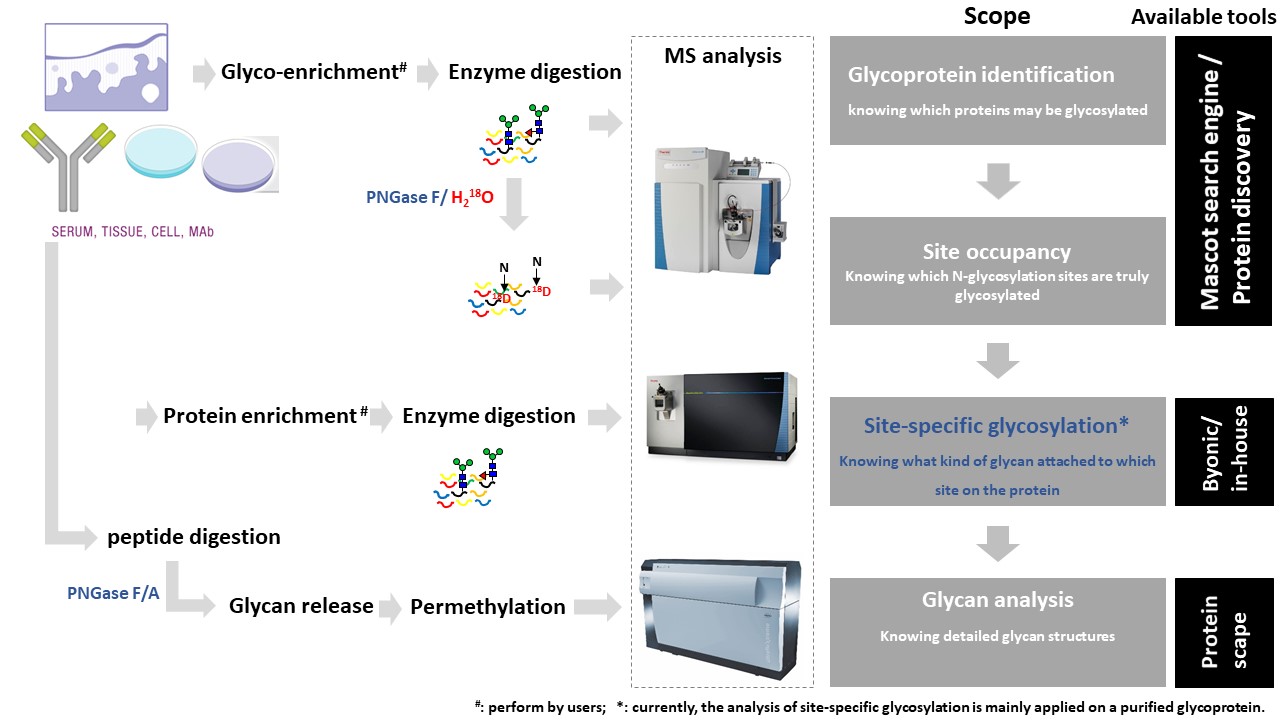
Glycopeptide analysis aims for N- and O-linked glycopeptide sequencing, indicating obtaining information from both glycans and peptides by mass spectrometry. This service allows to characterize glycoforms from purified proteins in solution form. The protein sequences will decide the digestion strategy and following analysis. The users are highly recommended to discuss with FGCZ experts before the submission of orders.Questions?
- Check our FAQs Glycan / Glycopeptide analysis page or contact us at proteomics at fgcz.ethz.ch
Workflow
Based on the biological question and on the type of sample, a typical workflow for processing the samples prior to MS and data analyses consists of:1. Protein digestion
The first step for the identification of unknown proteins is protein digestion using proteases (e.g. trypsin).Depending on the type of samples:
- Protein mixtures. Minimal recommended amount: 10 µg
2. LC-MS analysis
At FGCZ, the default LC-MS approach for protein identification is based on:- data acquisition on one of our Q-Exactive systems (Thermo, see Planetorbitrapusing Data Dependent Acquisition (DDA - also named Shotgun proteomics).
- In the case of experiments that require higher sensitivity, the data would be acquired on newer generation instruments such as the Orbitrap Lumos(Thermo)
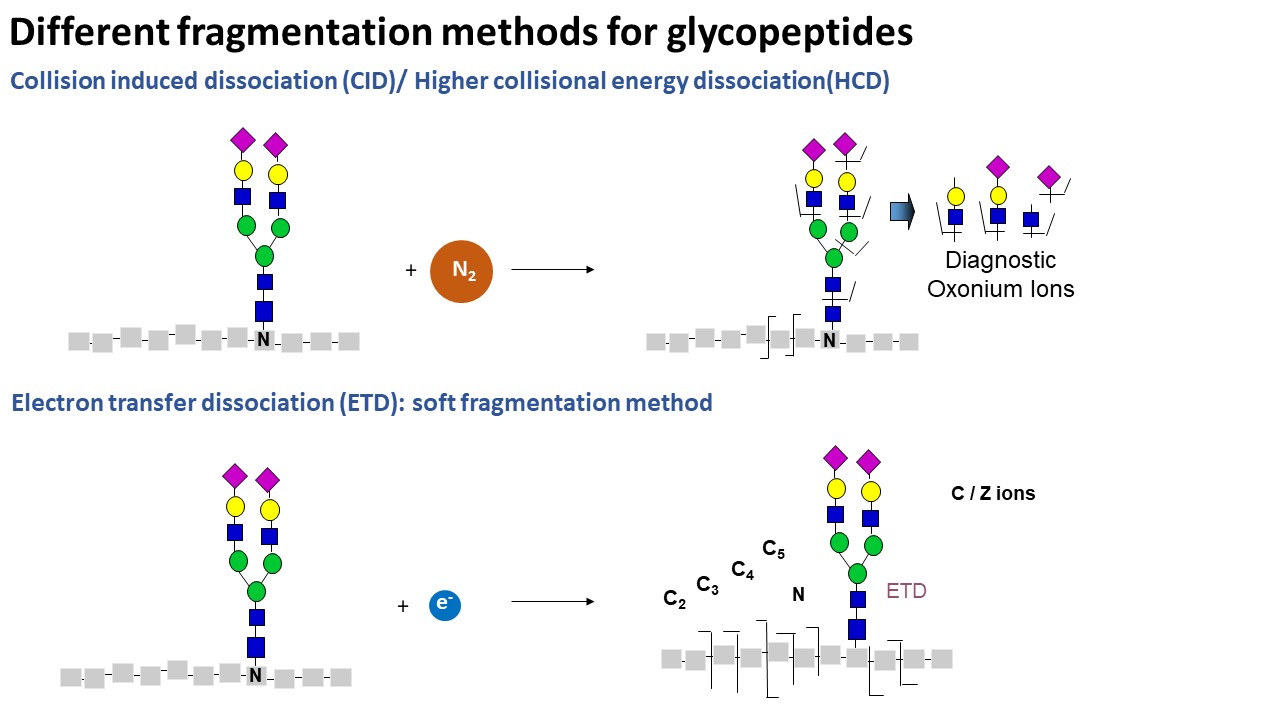
- For N-glycopeptide, the default method is HCD fragmentation. For O-glycopeptide, the default method is ETD fragmentation on Lumos MS.

- See all out instruments in our Instrument Intranet page)
3. Data analysis
To identify glycopeptide, the raw data will be searched against the whole database first and generate a focus database via Byonic search engine. The second search will be performed against the focus database with glycan database to save computation time.Some important points:
- for common organisms (e.g., human, mouse, ...) the databases are well-annotated and reviewed. We mainly use Uniprot databases.