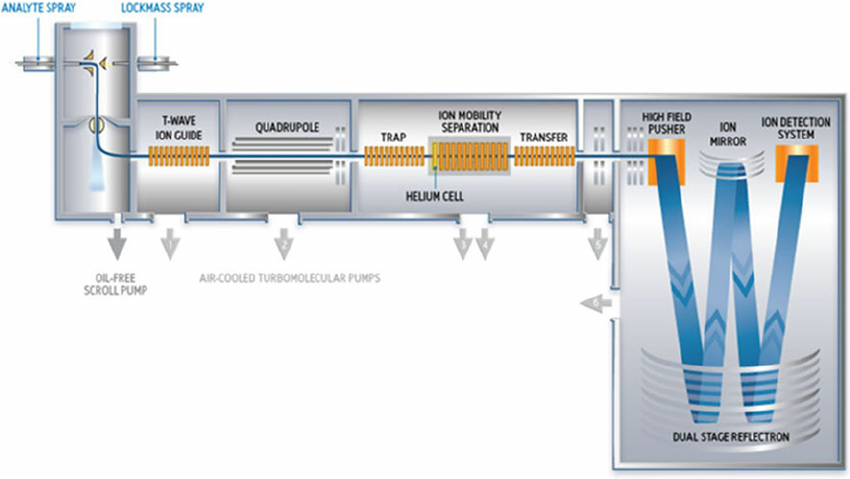
Proteomics > Our instruments
Please report errors and suggestions for improvements to Sibylle
If you have a running User Lab project and you would like to reserve an instrument, please check its availability on the FGCZ Instrument Reservation System (IRS), and submit a request for the desired time-slot. Your project coach, or the person responsible for the instrument, will contact you as soon as possible.
 .
.

The standard chromatographic separation is done with trapped elution prior the analytical elution in nano-flow mode. Separation is based on reversed-phase chromatography with C18.
Following trap and analytical columns are installed:
||
Table of contents
Please report errors and suggestions for improvements to Sibylle
Instrument Reservation
Instrument reservations can only be made by employees.If you have a running User Lab project and you would like to reserve an instrument, please check its availability on the FGCZ Instrument Reservation System (IRS), and submit a request for the desired time-slot. Your project coach, or the person responsible for the instrument, will contact you as soon as possible.
Proteomics equipment available at FGCZ
Default ESI and nLC setup
Ion source
For electrospray ionization (ESI), nearly all MS instruments are by default equipped with Digital PicoView nanospray sources (DPV)
DPV
Liquid chromatography
For sentitive, high peak capacity chromatography, our MS instruments are coupled to waters nanoACQUITY UPLC systems or ACQUITY UPLC M-Class systems.The standard chromatographic separation is done with trapped elution prior the analytical elution in nano-flow mode. Separation is based on reversed-phase chromatography with C18.
Following trap and analytical columns are installed:
- nanoEase M/Z Symmetry C18 100 A, 5um, 1/PK 180 um x 20 mm (Trap column)
- nanoEase M/Z HSS C18 T3 Col 100 A, 1.8 um, 1 P/K 75 um x 250 mm (Analytical column)
High-throughput chromatography
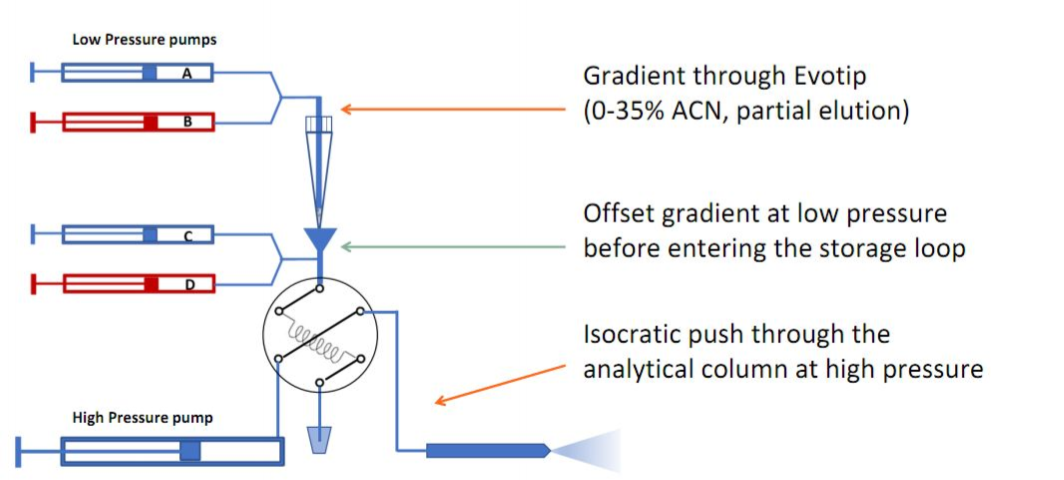
For large batches of samples, or in specific cases (e.g., very low-input material), the Evosep One platform is used for integrated purification and chromatographic separation. Evosep is particular suited for clinical proteomics (robust and fast).| Name | Detailed informations by clicking * | Principle * | Description | Remarks | Coupled to | Instruments at FGCZ | |
| EVOSEP ONE | High-throughput nanoUPLC system | Short gradients (i.e. 30-200 samples/day) for throughput. Samples are loaded on evotips and eluted directly from tips. Application | TIMSTOF_1,EXPLORIS_2, or LUMOS_2 | EVOSEP_1 and EVOSEP_2 | |||
MS Instruments
| Name | Detailed informations by clicking on the instrument * | Schema * | Mass analyzer | Fragmentation | Max resolution @ m/z 200 & Max scan rate | Operation Modes* | Instruments at FGCZ - Service/user lab access |
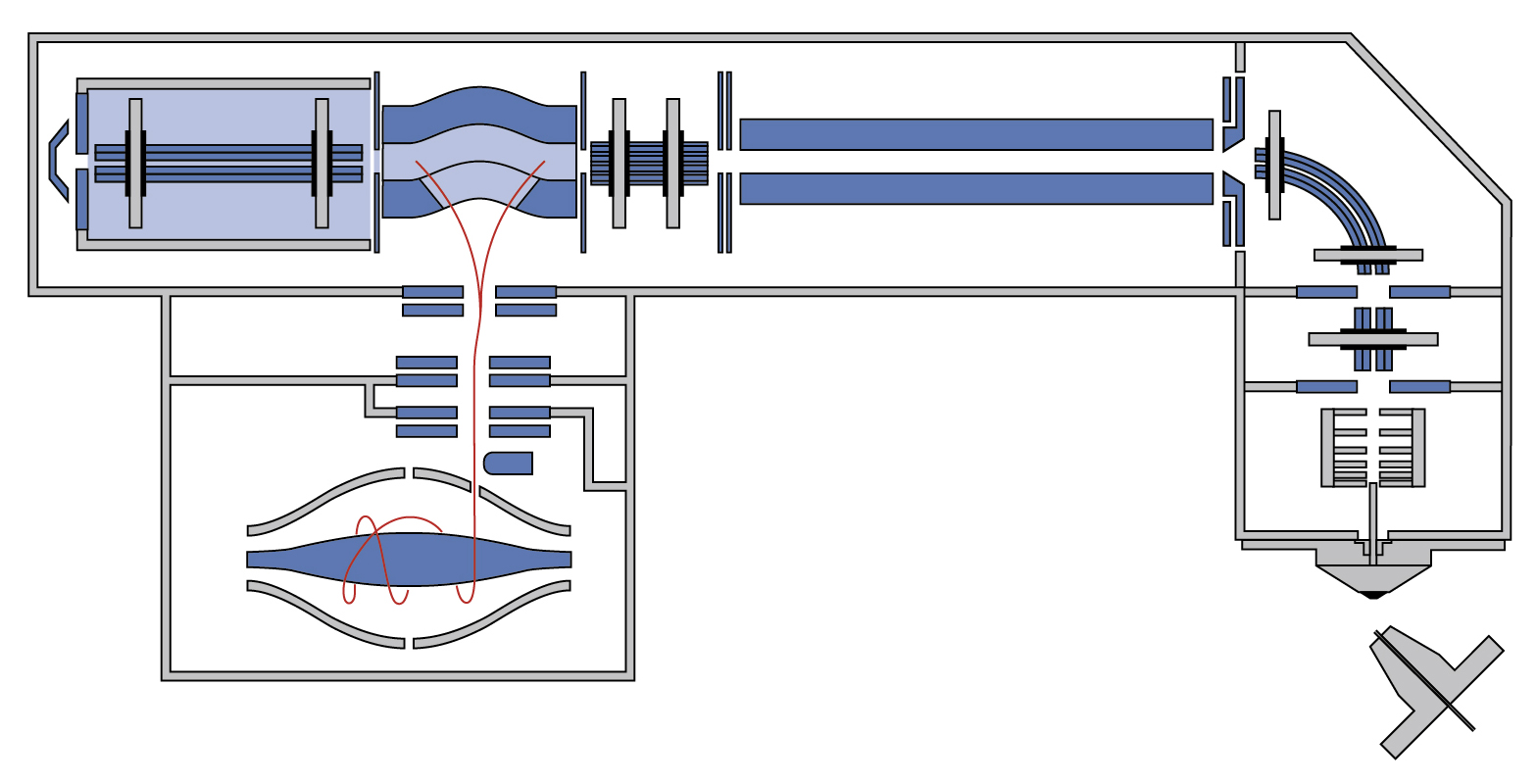
| Orbitrap Fusion Lumos | Tribrid™ combines quadrupole, Orbitrap, and linear ion trap | CID/HCD/ETD detected by the Ion Trap or Orbitrap at any level of MSn | 450,000 &30 Hz @ 7,500 resolution | DDA, Full MS, SIM, PRM, DIA, AIF, In-source fragmentation, Positive/negative ion switching, timed SIM, SPS-MS3, APD, FAIMS, Sure Quant | User LUMOS_1 and Service LUMOS_2 | ||
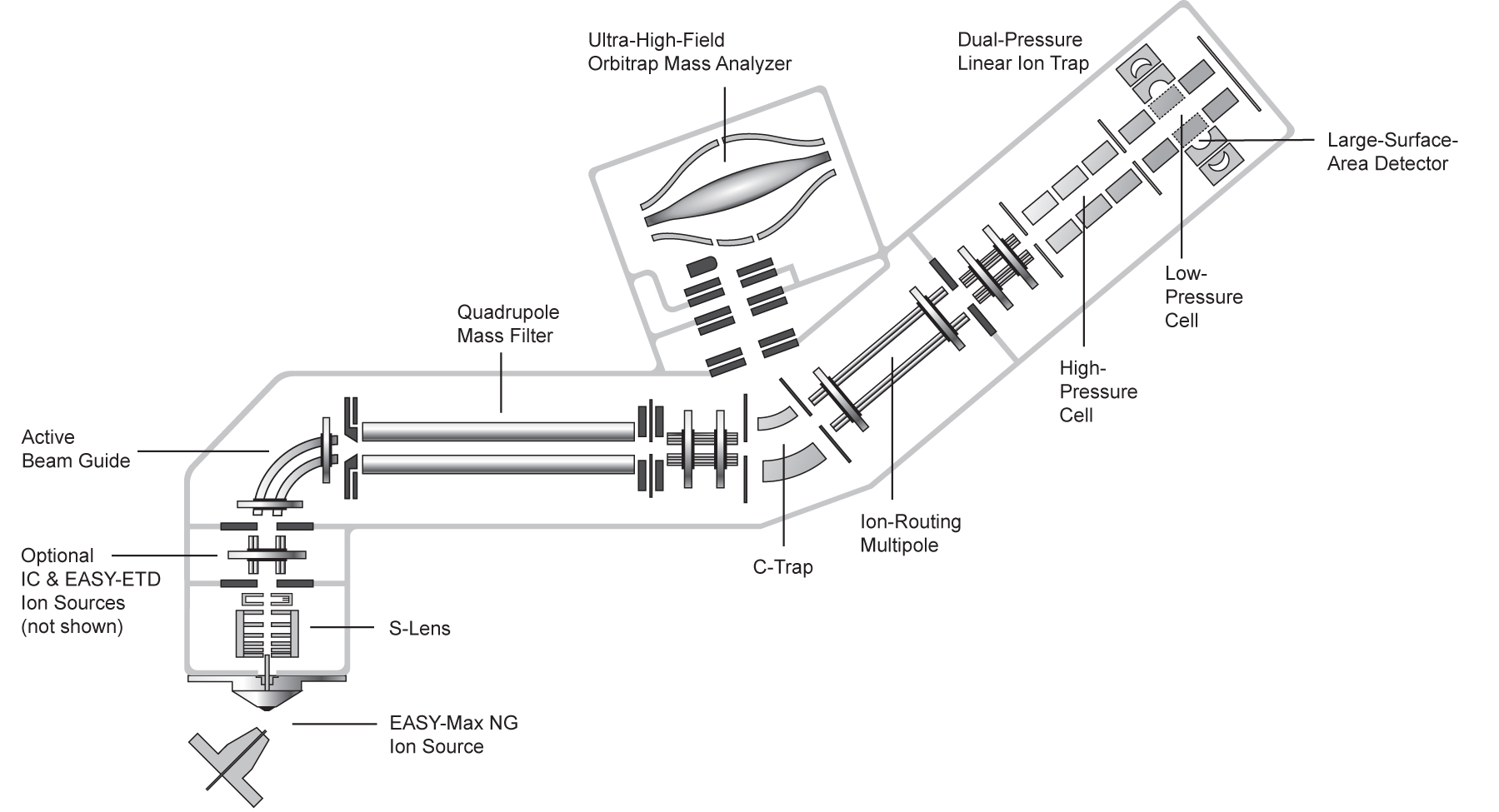
| Exploris 480 | Hybrid quadrupole, Orbitrap | HCD | 480,000 & 40 Hz @ 7,500 resolution | DDA, Full MS, SIM, PRM, DIA, AIF, In-source fragmentation, Positive/negative ion switching, timed SIM, APD, FAIMS, Sure Quant | User EXPLORIS_1 | ||
| Q Exactive | Hybrid quadrupole, Orbitrap | HCD | 140,000 & 12 Hz @ 17,500 resolution | DDA, Full MS, SIM, DIA, PRM | Service QEXACTIVE_1 and "test instrument" QEXACTIVE_2 (shared with metabolomics) | ||
| Orbitrap Fusion | Tribrid™ combines quadrupole, Orbitrap, and linear ion trap | CID/HCD/ETD detected by the Ion Trap or Orbitrap at any level of MSn | 450,000 &18 Hz @ 15,000 resolution | DDA, Full MS, SIM, PRM, DIA, AIF, In-source fragmentation, Positive/negative ion switching, timed SIM, SPS-MS3, FAIMS | User FUSION_2 | ||
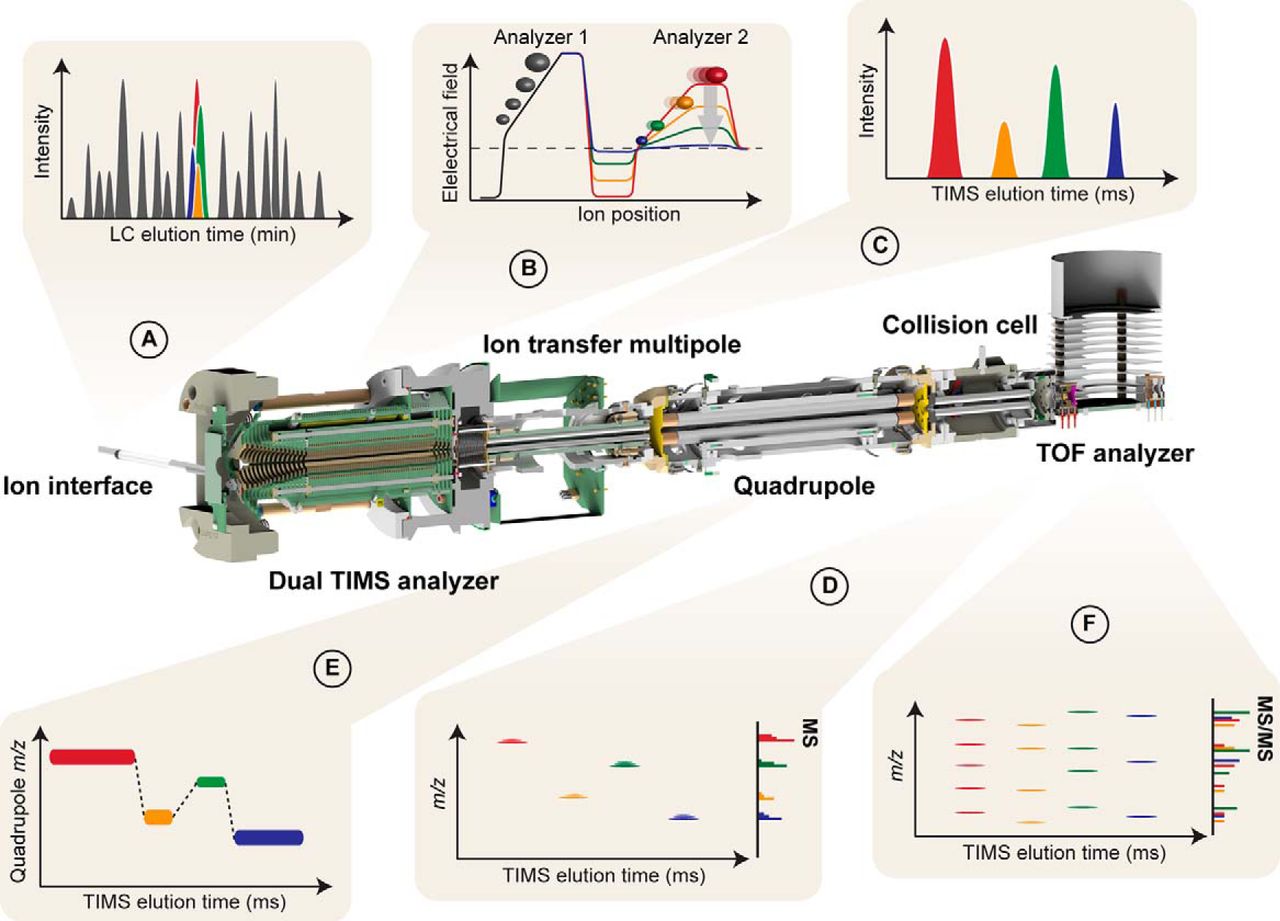
| timsTOF Pro | https://doi.org/10.1074/mcp.TIR118.000900 | dual Trapped ion mobility (TIMS), quadrupole, time-of-flight (TOF) | CID | >200 for resolution of ion mobility, >100 Hz for PASEF | Parallel Accumulation Serial Fragmentation (PASEF), diaPASEF | User TIMSTOF_1 | |
| timsTOF Flex MALDI-2 | See details here | MALDI-2 post-ionization, dual Trapped ion mobility (TIMS), quadrupole, time-of-flight (TOF) | CID | >200 for resolution of ion mobility, >150 Hz for PASEF | From 4D-Omics to molecular imaging without stopping, changing, modifying or compromises (learn more), diaPASEF | Service timsTOF_Flex_1 | |
| RapiFlex MALDI TOF-TOF | Matrix Assisted Laser Desorption Ionization — Time of Flight Mass Spectrometry (MALDI-TOF-TOF) | high energy collision dissociation MS/MS techniques and in-source decay in the reflector mode of the mass analyzer (reISD) as a pseudo-MS/MS technique | ≥ 50,000 for Somatostatin 28 (m/z 3,147.5) FWHM | MS in liner or reflectron mode, HCD-MS/MS, Imagining (Imaging Pixel shape Squared pixel for best tissue coverage with variable pixel size of 20 μm to 100 μm; Imaging Speed Up to 40 pixel / sec. | Service RapiFlex_1 | ||
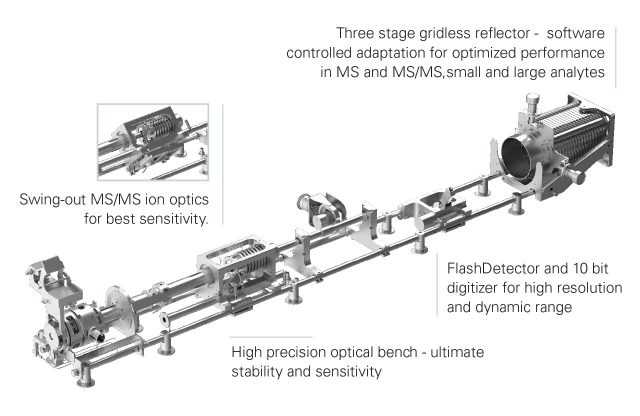
| Q-TOF SYNAPT G2-Si | Waters Synapt G2-SI 8k with ETD, or 4k with IMS | CID and ETD | Up to 60,000 | DDA, Full MS, DIA | Service G2SI_1 and G2SI_2 | ||
| * • Full MS with high-resolution accurate mass detection • Selected Ion Monitoring (SIM) with high-resolution accurate-mass detection • Parallel Reaction Monitoring (PRM) for improved screening and quantitative confidence • Data-Independent Acquisition (DIA) for unbiased identification and quantitative analysis • All-Ion Fragmentation (AIF) in the HCD collision cell with high-resolution accurate-mass detection • In-source fragmentation of all ions • Positive/negative ion switching on chromatographic timescale • On-the-fly data-dependent decision making ddMS2 > data dependent acqusition (DDA) • Timed SIM for scheduled data acquisition of the targets of interest • synchronous precursor selection (SPS)-based MS3 • advanced peak determination (APD) algorithm • High-Field Asymmetric waveform ion mobility (FAIMS) • laser-induced dissociation (LID) | |||||||
Other detectors
| Name | Detailed informations by clicking on the instrument * | Schema * | Description | Operation Modes* | Instruments at FGCZ - Service/user lab access | ||
| ACQUITY QDa Mass Detector | Single Quadrupole Mass Detection | ACQUITY QDa is as intuitive as an optical detector. Integrated in Acquity UPLC H-Class Plus | Service QDA_1 | ||||
| ACQUITY FLR Detector |
| 2475 Fluorescence (FLR) Detector | The detector helps achieve high-sensitivity and low-dispersion methods. Integrated in Acquity | Service Characterization_Nlinked_glycans | |||
Other equipment for MS workflow
Sample preparation
| Name | Detailed informations by clicking * | Principle * | Description | Remarks | Coupled to | Instruments at FGCZ | |
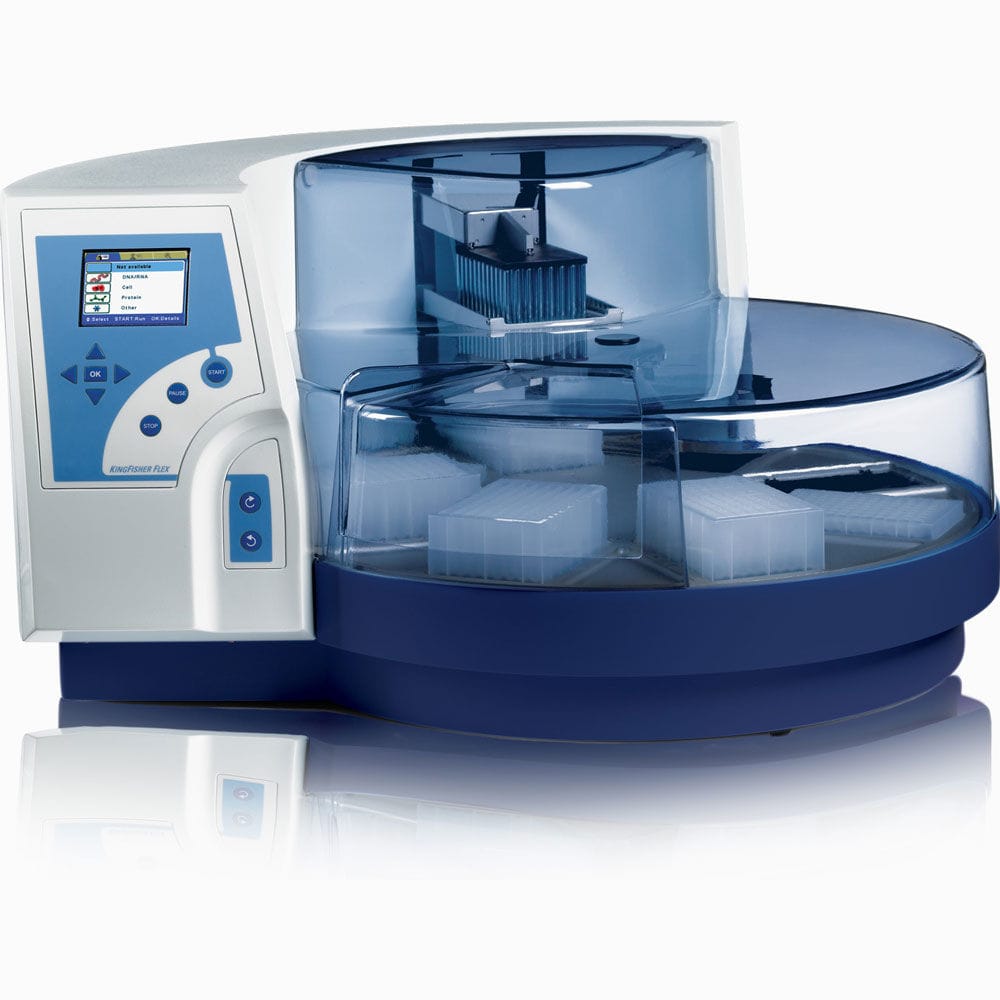
| KingFisher™ Flex | Automated sample preparation system. Process up to 96 samples simultaneously | Prepare samples efficiently from a variety of materials. It offers highly versatile, automated magnetic-particle processing for DNA/RNA, protein or cell purification from virtually any source. Protocol Automated Magnetic Separations for Proteomics | NA | KingFisher Flex | |||
| HTX M5 Sprayer | Te HTX M5 Sprayer™ is an easy-to-use, versatile spraying system that provides automated process for Sample Preparation in Mass Spectrometry Imaging. | Proprietary technology providing very small matrix droplets ( <5 microns), High fow rate and fast sample prep (2 to 18 minutes per slide), Highly consistent matrix deposition across entire sample area (+/- 3% by weight), Unique use of temperature and nitrogen flow to control evaporation rate and matrix crystal formation | NA | HTX M5 sprayer | |||
| Barocycler NEP2320 Enhanced | Pressure Cycling Technology (PCT). Sample Preparation System - Process up to 16 samples simultaneously | Pressure Cycling Technology (PCT) is a unique, patented technology platform based on | NA | Barocycler (PCT) | |||
| Covaris LE220 | Ultrasonicator for DNA shearing and tissue lysis 1 to 8 tubes at a time parallel processing; 96 tube capacity | Accurate, reproducible, and robust sample preparation powered by Adaptive Focused Acoustics® (AFA®) technology. | NA | GEML - Covaris LE220 | |||
Sample fractionation
| Name | Detailed informations by clicking * | Principle * | Description | Remarks | Coupled to | Instruments at FGCZ | |
| Agilent HPLC offline | Analytical-Scale LC Purification System for offline prefractionation | Analytical-scale purification LC system ideal for purification of multi-milligram quantities of materials. Dynamic flow range up to 10 mL/min at maximum 600 bar. Collection of up to 4 x 96 fractions in microtiter plates | NA | LC1100 and LC1200 | |||
| FAIMS Pro online | Post-ionization gas phase fractionation based on differential ion mobility - space separation. It offers separation based on a combination of factors, such as charge state, shape, conformation, and size of gas phase ions. | The FAIMS Pro interface reduces chemical noise and matrix interferences, resulting in improved assay robustness and increased sensitivity. It can be particularly beneficial in some proteomics applications.Details FAIMS Pro & FAIMS-MS2 for TMT QuantificationShotgun ProteomicsFAIMS Exploris 480 | EXPLORIS_1 | NA | |||
Commercial softwares for data analysis
| Name | Detailed Informations * | Use | Need help? Contact person at FGCZ | Software access at FGCZ | |||
| Proteome discoverer | PD commercial software offers a full suite of analysis tools with the flexibility to address multiple research workflows, and an easy-to-use, wizard·driven interface. Supporting multiple database search algorithms (SEQUEST HT, Mascot, Byonic, MS Amanda, and ProSightPD) and multiple dissociation techniques (CID, HCD, ETD, and EThcD) | wiki | pd @ fgcz-rds-2 | ||||
| Spectronaut | Commercial discovery proteomics software for the targeted analysis of DIA measurements from various MS platforms. | wiki or Jonas | Spectronaut | ||||
| SpectroDive | Commercial Targeted proteomics software for the analysis of PRM and MRM measurements from various MS platforms | Jonas | SpectroDive | ||||
| Byonic™ | Commercial software with top-down and bottom-up search capabilities. | Chia-wei | Byonic @ FGCZ-h-076 | ||||
| ScaffoldPTM | Visualize and Quantitate Site Localized PTMs from various search engines | Jonas | ScaffoldPTM | ||||
| ScaffoldQ+S | Visualize and Quantitate iTRAQ/TMT/SILAC from various search engines | Jonas | ScaffoldQ+S | ||||
| IntactMass™ | Commercial software for mass spectra of intact (undigested) proteins. | Serge | INTERNAL USE ONLY - IntactMass @ FGCZ-h-078 | ||||
| Progenesis QI for Proteomics | Proteomics software for LC-MS data | Jonas | ProgenesisQI_Proteomics@170 or DISCONTINUED - used only in special cases ProgenesisQI for Proteomics on 166 | ||||
| ProteinPilot™ | ProteinPilot™ Software is a paradigm shift in protein identification and relative protein expression analysis for protein research. Combining sophisticated algorithms with an intuitive interface, you can now confidently identify more proteins and search large numbers of post translational modifications, without increasing search time or false positives. | Jonas | DISCONTINUED - ProteinPilot V4 | ||||


.jpg)

.jpg)