Ion Torrent PGM
The Ion Torrent website contains the latest information on semiconductor sequencing technology.
The newest available protocols and other information are available at the Ion Community website (email registration required).The following Decision Tree graph may be helpful for planning a sequencing experiment on the PGM: DNA sequencing on PGM Decision Tree. Nevertheless, get in contact with us before preparing any sequencing experiment on the PGM:sequencing@fgcz.ethz.ch
| 
| 
| |
| Chip Type | 314v2 | 316v2 | 318v2
|
| Read Length | up to 400 bp | up to 400 bp | up to 400 bp
|
| Million wells per chip | 1 | 6 | 11
|
| Number of reads (millions) | 0.4 - 0.5 | 1.9 - 2.5 | 3.3 - 4.4
|
| Throughput * for 200-base reads | 30 -50 Mb | 300 - 600 Mb | 0.6 - 1 Gb
|
| Throughput * for 400-base reads | 60 - 100 Mb | 0.6 -1 Gb | 1.2 - 2 Gb
|
| Accuracy | >99.99% consensus accuracy and >99.5% raw accuracy
| ||
| Available at FGCZ | yes | yes | yes
|
| * Expected throughput with > 99% aligned/measured accuracy | |||
Principles of library preparation for Ion Torrent sequencing
The library preparation procedure via ligation is illustrated here: 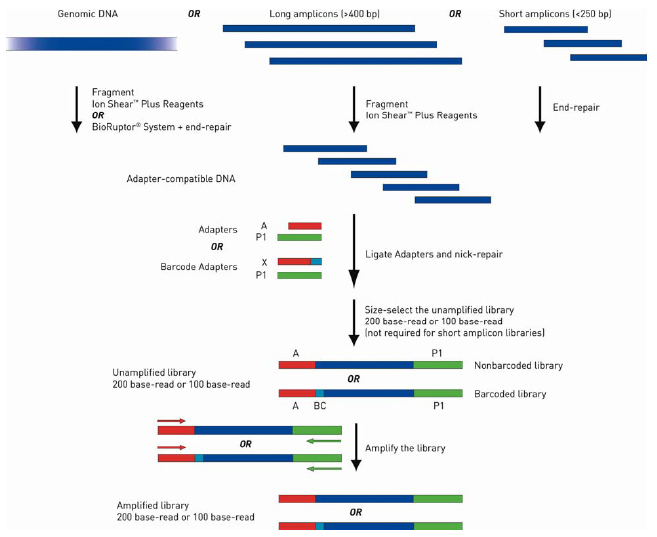
| First, genomic DNA (gDNA) or pooled long (>400 bp) amplicons are fragmented to appropriately sized, blunt ended DNA fragments that are ready for ligation to Ion adapters. Amplicons <250 bp in length (short amplicons) do not require fragmentation but do require end repair for ligation. Next, the adapter ready DNA is ligated to Ion compatible adapters, followed by nick repair to complete the linkage between adapters and DNA inserts. If barcoded libraries are required, substitute adapters from the Ion Xpress Barcode Adapters 1–96 kits at this step. The adapter ligated library is then size selected for optimum length according to target read length. Short amplicon libraries do not require size selection, but the amplicons should be designed to be shorter than the median insert size for the target read length of the library. Final amplification of the library is optional, depending on the amount of input DNA and your experimental requirements. There are also available Ion AmpliSeq DNA or RNA Panels (Ready-to-Use and Custom) for targeted library preparation. Please check http://www.ampliseq.com. The newest available protocols and other information are available at the Ion Community website (email registration required). | ||
| Amplicon libraries can be also prepared directly using Fusion Primers method. Here is the list of 96 barcodes which can be used for primer design. The size of libraries/amplicons matters a lot in the ePCR step. The amplicon preparation should be done strictly following the protocol. The newest available protocols and other information are available at the Ion Community website (email registration required). | |||
Input material requirements
gDNA sequencing
- 100 ng or 1 ug genomic DNA for fragmenting enzymatically with the Ion Shear Reagents.
RNA sequencing
- 1–500 ng of poly(A) RNA, or 25–500 ng of rRNA‐depleted total RNA or high-quality total RNA