Proteomics > Which service should I request? > Characterization of Purified Biomolecules > disulfide bond assignment
Disulfide bond assignment
Table of contents
General description
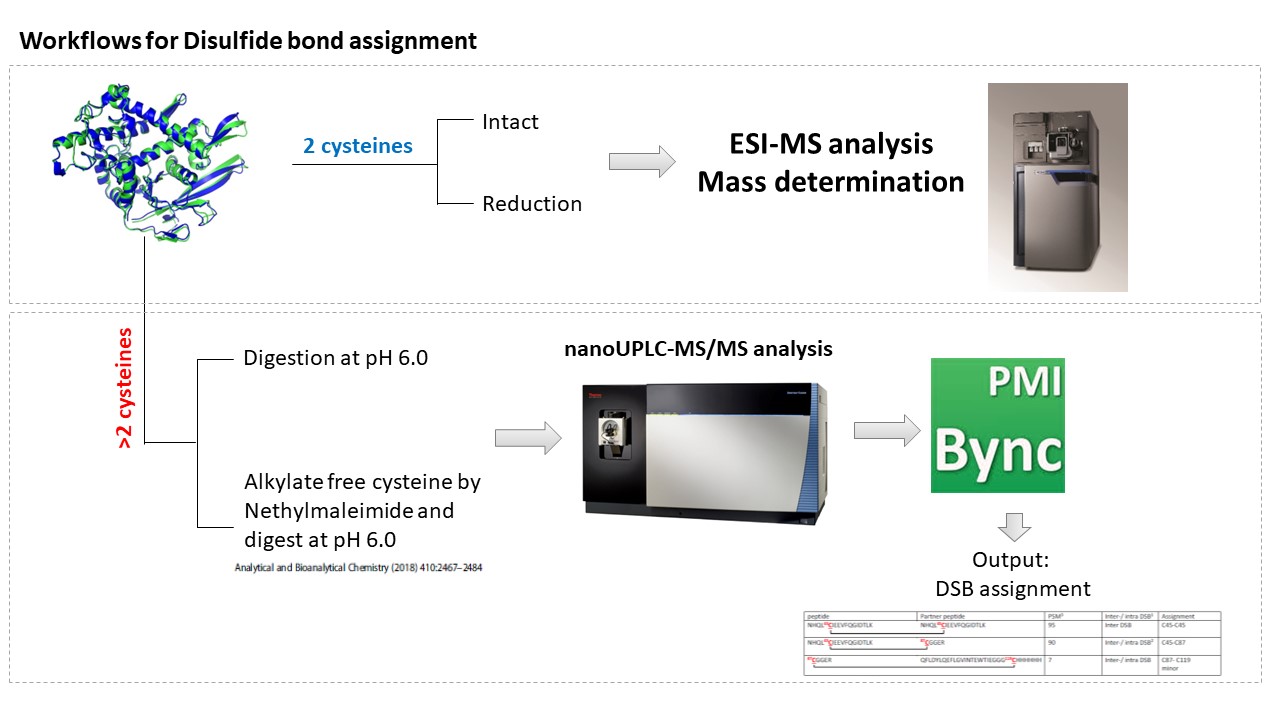
The linkages of the disulfide bonds (DSB) play important roles in protein stability and activity. The Mass spectrometry-based (MS-based) technique is one of the accepted tools for DSB analysis. FGCZ has two strategies for disulfide bond analysis.Workflow
- For proteins containing 2 cysteines AND form monomer: The intact molecular weight of proteins before and after reduction will be measured by Waters SYNAPT G2 mass spectrometer. Based on the mass shift, the status of two cysteines will be shown.
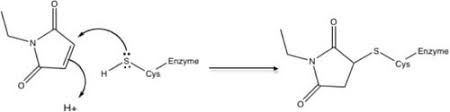
- For proteins containing more than 2 cysteines: To confirm the purity and formation of the proteins, intact mass determination is required before digestion. This can either be performed through FGCZ service Protein / peptide mass determination or the data provided by the users. The free cysteine(s) will be first blocked with or without 5mM N-ethylmaleimide (NEM) for 30 mins. Then, the proteins will be digested by the enzyme in acidic conditions and incubated for a few hours to overnight. The digested peptides will be analyzed by Thermo FUSION Orbitrap mass spectrometer and the data will be analyzed by Byonic (Protein Metrics).
Requirements and considerations
- volume: > 5 uL
- minimal amount: 1-5 ug; optimal amount: 10 ug
- dissolve samples in volatile solvents or free of non-volatile buffers, salts, detergents, and primary amine
Reference
- Sung, Q.C. et al 2016, Biochimica et Biophyica Acta 2016. 1188-1194 Evaluation of disulfide scrambling during the enzymatic digestion of bevacizumab at various pH values using mass spectrometry))
- Na, S et al 2015, 11, 1156-1164 Characterization of disulfide bonds by planned digestion and tandem mass spectrometry