Proteomics > Which service should I request? > Proteome Identification and Quantification > Targeted Quantification
Some examples of applications are:
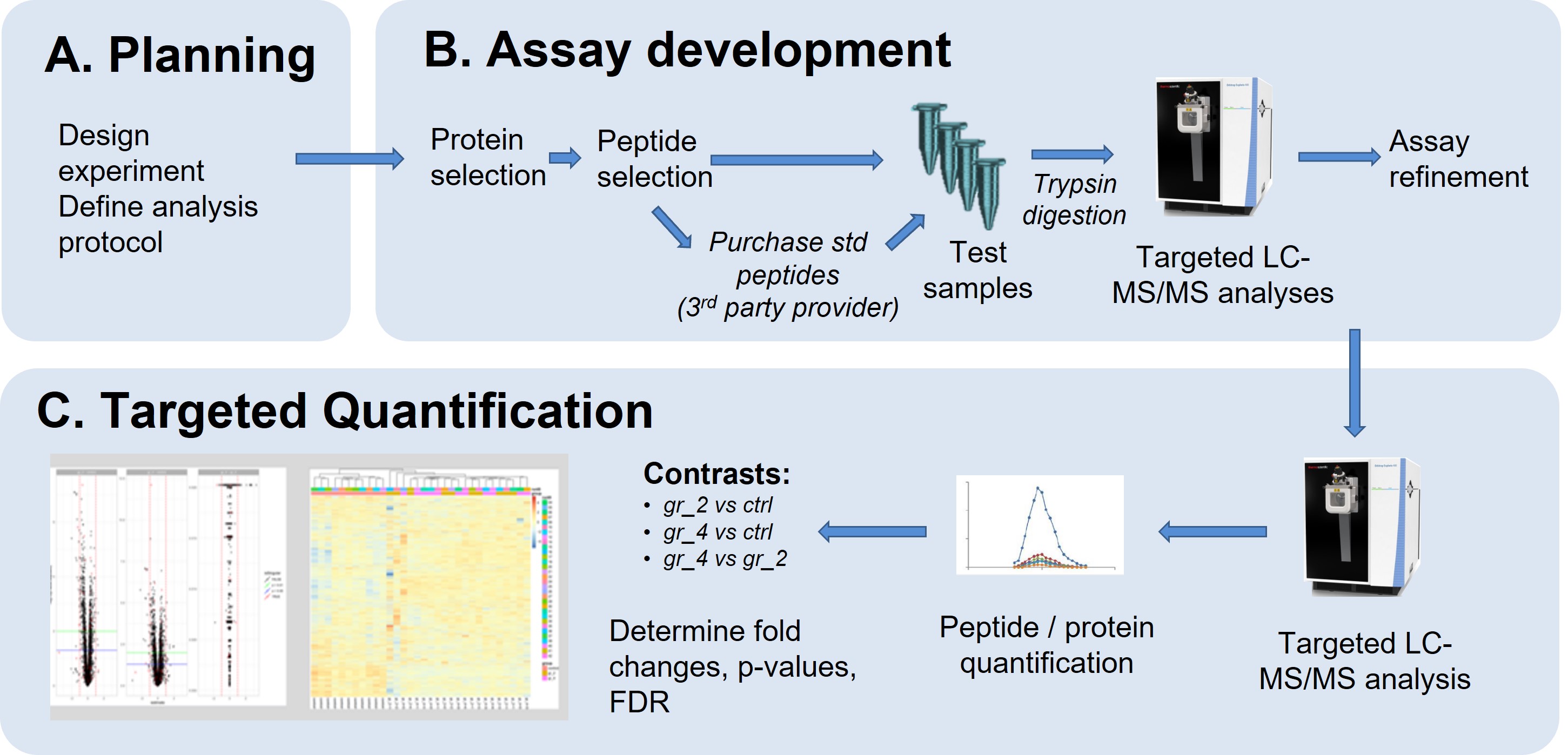
All targeted quantification methods consists of 3 steps:
Please contact us at proteomics at fgcz.ethz.ch for a meeting, or directly contact your coach If you already have a B-Fabric project at FGCZ.
Goal:
Targeted Quantification
Table of contents
General description
Mass spectrometry-based targeted quantification methods aim at the detection of changes of protein abundances in biological samples.Some examples of applications are:
- The measurement of protein abundances in biological pathways across multiple samples in different perturbation states
- The verification of a panel of bio-markers with potential impact to e.g. cancer diagnosis and prognosis
- The analysis of "the activity status" of pathways and protein complexes for example by determining the phosphorylation stoichiometry or protein subunit stoichiometry
Workflow
Selected/Parallel Reaction Monitoring (SRM/PRM) and Data Independent Acquisition (DIA) based methods allow the relative or absolute quantification of a preselected set of proteins across multiple conditions with high reproducibility and sensitivity. The FGCZ is mostly using PRM as well as SureQuant™, an internal standard triggered targeted quantification (IS-PRM) workflow that is dynamically controlled and independent of time scheduling.All targeted quantification methods consists of 3 steps:

A. Planning
As a first step, we need to clarify the goal of the project and clarify the analytic and bioinformatics strategy. A proper experimental design will be defined together with FGCZ staff.Please contact us at proteomics at fgcz.ethz.ch for a meeting, or directly contact your coach If you already have a B-Fabric project at FGCZ.
B. Assay development
In targeted quantification experiments, the development of the assay is often the most time-consuming and challenging step.Goal:
- define which peptides are unique for the proteins of interest (proteotypic peptides)
- select the best peptides for each protein (ideally >=3 peptides/protein)
- purchase the isotopically labelled standard peptide (from 3rd party companies, i.e. JPT)
- test the workflow with some samples (protein digestion, spike of std. peptides, SureQuant Method, LC-MS/MS analysis)
- assay refinement
- assay finalization
C. Targeted quantification
Once the assay has been developed, it can be applied on samples on almost any number of samples. Typical experiments analyse dozens of samples.Requirements and considerations
- The assay development is mandatory, and it can require many weeks of time just for the delivery of the standard peptides
- A typical experiment focuses on ~10 proteins, for up to 25-30 peptides. The analysis of an higher number of proteins is possible.
- Replicates: 4+ for in-vitro experiments, 5/10+ for animal/human experiments (depending on the results of the sample size estimation)
- Buffer composition and turnaround time: every experiment requires some optimization, please contact us at proteomics at fgcz.ethz.ch
Ready-made protein panels
Vendors often provide ready-made protein panels for targeted quantification. Some examples:[+] AKT pathway, AKT (phospho), AKT (isoform), RAS (isoform), TP53 - Thermo Scientific
[+] Cytokines (human) - JPT
[+] 500+ plasma/serum proteins (human, PQ500™) - Biognosys
Data acquisition and data analysis
- The data can be acquired and analysed in multiple ways, but SureQuant is the most common strategy used at FGCZ (see below).
- after data acquisition, the analysis is performed using Skyline or SpectroDive (Biognosys)
- Statistical analysis is performed using our published prolfqua package (incl. MSStats)