Proteomics > Which service should I request? > Glycan / Glycoprotein analysis > Characterization of N-linked glycans
Questions?
Characterization of N-linked glycans
Table of contents

General description
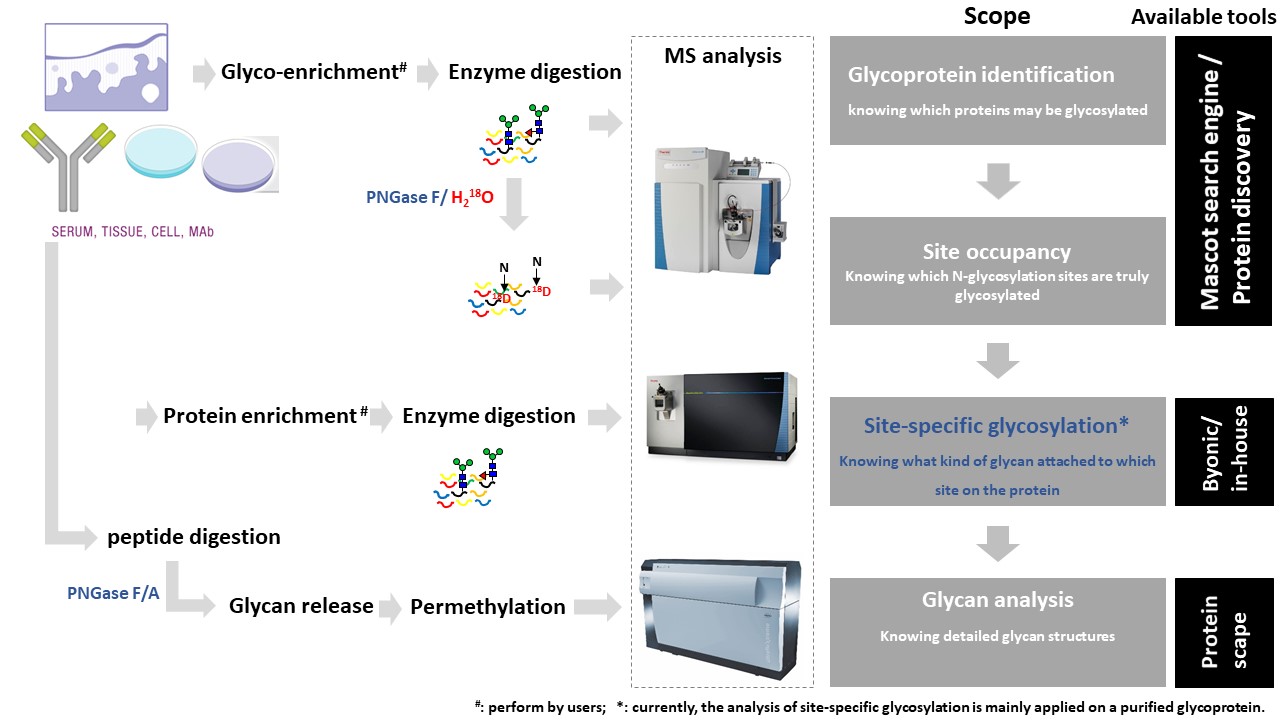
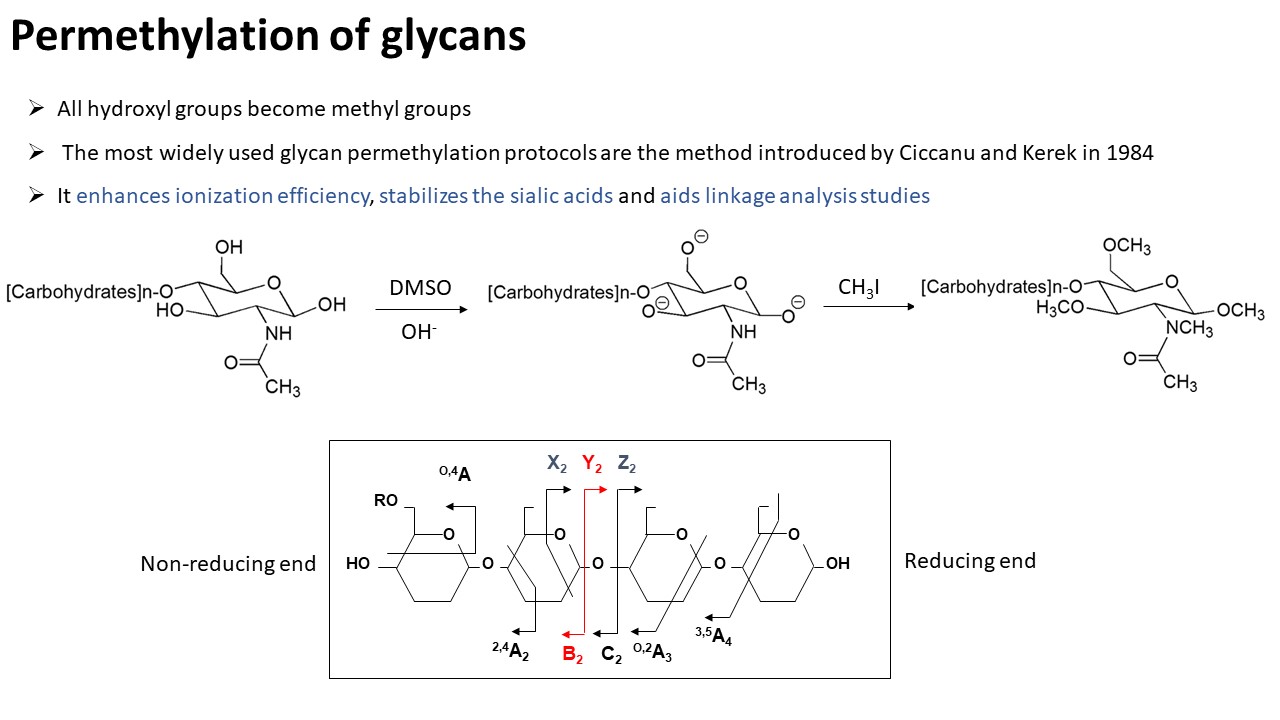
Glycomics "is the systematic study of all glycan structures of a given cell type or organism" and is a subset of glycobiology. To characterize N-glycans, the protein mixtures would be first extracted from their biological resources. Then, the N-glycans are released by PNGase F or PNGase A depending on the species. To increase the sensitivity of glycans on MS-based detection, glycans would be further permethylated before MS analysis. The MS profile can provide an overview of N-glycome from samples. For advanced structure characterization, tandem mass spectrometry would be applied to confirm the linkage of glycan structures. For O-glycomics, please contact us for further information.Questions?
- Check our FAQs Glycan / Glycopeptide analysis page or contact us at proteomics@fgcz.ethz.ch
Workflow
Based on the biological question and on the type of sample, a typical workflow for processing the samples prior to MS and data analyses consists of:1. Protein digestion/ glycan release and permethylation
- Protein digestion: Proteins mixtures extracted from different biological resources, for example, cells or biological fluid, are first digested into peptides.
- Glycan release: For N-glycomic analysis, samples are treated with PNGase F/A. The released glycans would be purified by a reverse-phase cartridge.
- Permethylation: Glycans are chemically methylated by methyl iodide.
