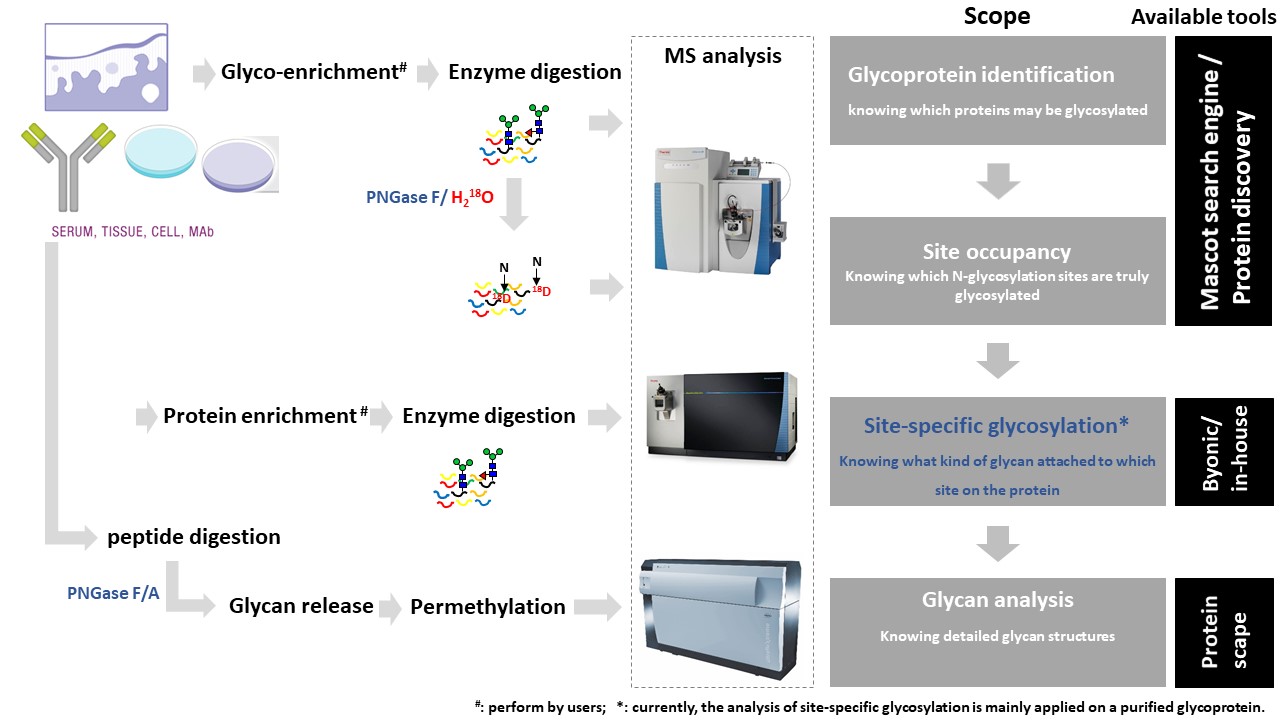
Proteomics > Which service should I request? > Glycan / Glycoprotein analysis > N-linked glycans site occupancy
Questions?
Examples of instruments are the Q-Exactive (Thermo) or, in case of experiments that require higher sensitivity, Orbitrap Lumos(Thermo)
See all out instruments in our Instrument Intranet page)
The peptides are considered as glycopeptides with the occupied site when the Asn (N) of the consensus site N-X-S/T (X≠P) is identified as deamidated to Asp (D).
N-linked glycans site occupancy
Table of contents

General description
Each glycoprotein may contain a number of potentially N-glycosylated sites, each of which may or may not be glycosylated. The scope of site-occupancy analysis is to identify in a complex protein mixture the peptides which are truly glycosylated.Questions?
- Check our FAQs Glycan / Glycopeptide analysis page or contact us at proteomics at fgcz.ethz.ch
Workflow
N-linked glycans can be enzymatically released from peptides and the original N-glycosylation sites would be converted to Asp (D), resulting in a 1 Dalton increment compared to their unglycosylated congeners. However, spontaneous deamination occurs frequently in nature. In order to differentiate spontaneous deamination and enzymatic release, the reaction can be performed in the presence of heavy water (H218O), thus increasing the mass difference to 3 Dalton. The sites which are truly glycosylated can be identified based on the conversion of Asn (N) to Asp (D). The approach can be used in combination with quantitative strategies.1. Glycoprotein enrichment
Enrichment of glycoprotein from a protein pool using lectins or antibodies (performed by the customer)2. N-Glycan release
Release of N-Glycans using PNGase F or A enzymes. Optionally the reaction can be performed in H218O water, but this is not the standard protocol at FGCZ3. Protein digestion
Protein digestion using proteases (e.g. trypsin). In most cases the standard protein identification workflow is applied3. LC-MS analysis
At FGCZ, the default LC-MS approach for protein identification includes data acquisition on one of our LC-MS/MS systems using Data Dependent Acquisition (DDA - also named Shotgun proteomics).Examples of instruments are the Q-Exactive (Thermo) or, in case of experiments that require higher sensitivity, Orbitrap Lumos(Thermo)
See all out instruments in our Instrument Intranet page)
4. Data analysis
Peptide and protein identification using search engines (e.g. Mascot) and protein sequence databases. For common organisms (e.g., human, mouse, ...) the databases are well-annotated and reviewed. We mainly use Uniprot databases.The peptides are considered as glycopeptides with the occupied site when the Asn (N) of the consensus site N-X-S/T (X≠P) is identified as deamidated to Asp (D).