Proteomics > Which service should I request? > Proteome Identification and Quantification > Proteome Quantification - TMT
Depending on the experimental design and the biological question, we also offer Label-free proteome quantification methods
Thermal Proteome Profiling (login required to access this page)
Proteome Quantification - TMT
Table of contents
General Description
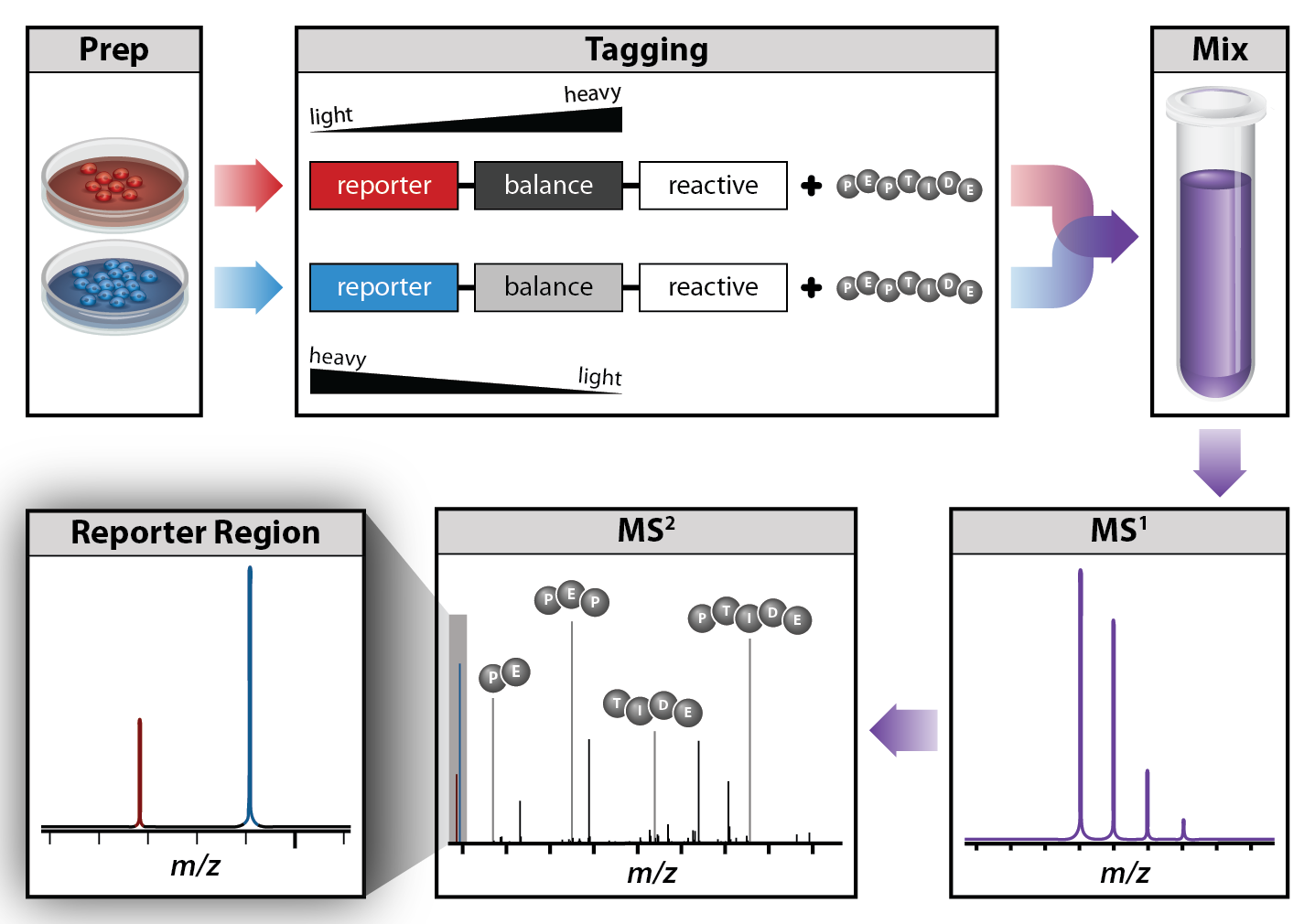
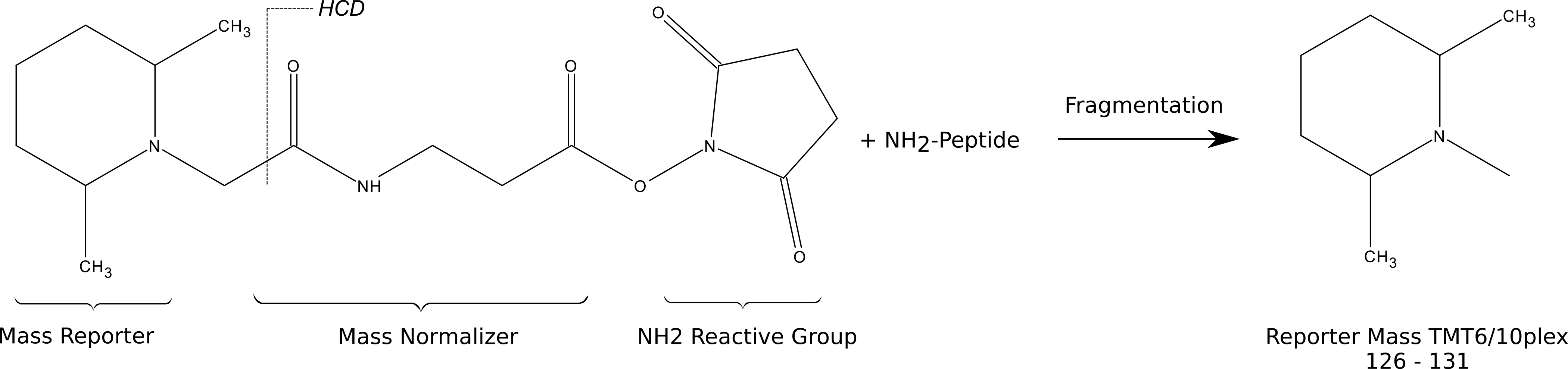
Tandem mass tags (TMT) are isobaric reagents that provide multiplexing capabilities for relative quantitative proteomics analysis.- The tags consist of three parts with varying degrees of isotope (C15, N13) incorporation along the mass balancing and reporter groups such that lighter reporter regions are paired with heavier balance regions and vice versa.
- Adding the tags to peptides results in the same mass shift for peptides from different conditions.
- After pooling the samples, peptides from different conditions appear as a single precursor in MS1.
- Upon fragmentation, the unique reporter ions used for relative quantification are released in addition to normal peptide fragment ions used for peptide identification.
Depending on the experimental design and the biological question, we also offer Label-free proteome quantification methods
Why multiplexing?
This system allows to multiplex peptides from up to 18 samples and comes with several advantages over label-free or metabolic labeling (SILAC) approaches.- Reduced MS1 complexity compared to SILAC
- Increased throughput
- Reduced analysis time
- Fewer missing values (compared to MS1-based LFQ)
- Smaller variances
- Reduced input requirement (important for e.g. PTM analyses such as phosphoproteomics)
- Higher precision compared to MS1-based LFQ
- Not dependent on sample origin (unlike SILAC)
- Potential for significantly increasing analytical depths when combined with fractionation approaches (see below)
Important points to consider
- Protein identification rate is lower compared to LFQ-approaches due to more stringent settings during data acquisition and data analysis (see fractionation for more info)
- Multiplexing is not ideal for the analysis of samples with large differences in protein dynamic range, complexity and composition (e.g. multiplexing of serum and tissue samples)
- Ratio compression can lead to smaller, potentially non-significant fold-changes especially for peptides with low reporter ion signals
- Near complete quantitative matrix in combination with ratio compression can produce artefacts of apparent presence of peptide in a sample when it should not be present
- Peptide retention time, ionization, "flyability" and fragmentation efficiency can be affected by the added on tag
Default Workflow
Protein digestion, peptide labeling and multiplexing
- Protein digestion is most commonly done using an SP3-based method
- Alternative digestion protocols can be used (FASP, TCA precipitation, PCT, on-bead) as long as the final buffer components (volatile, no primary amines) are compatible with the labeling reaction (see below)
Sample fractionation - OPTIONAL
- In order to obtain deeper proteome coverage a first dimension separation (e.g. HPLC, SPE, HILIC, IEF, SEC, gel-based) can be introduced before loading samples onto the nano-LC system
- Multidimensional liquid chromatography provides the most extensive proteome coverage by efficiently reducing sample complexity through a combination of offline and online fractionation
- The following methods are established and routinely used at the FGCZ:
- High-pH offline fractionation
- SPE-based methods such as stage-tips or Sep-pak
- Zwitterionic (HILIC) magnetic beads (automated)
Data acquisition
MS2-based Data-dependent acquisition- Samples are analyzed on a latest-generation mass spectrometer (e.g. Thermo Exploris 480 or Orbitrap Fusion Lumos)
- Quantification is routinely done by extracting reporter ion intensities from MS2 spectra
- Upon request we can acquire reporter ion intensities in MS3 mode on am Orbitrap Fusion Lumos
Data analysis
- Our current standard workflow includes the analysis using Fragpipe 19.1
- Downstream analysis is similar to LFQ-based analysis
- Proteome Discoverer 2.5 (Thermo) in combination with Sequest or Mascot can be done upon request
Requirements and Considerations
- Current TMT kits allow multiplexing of 6, 10, 11, 16 or 18 samples. As we charge one complete kit, we recommend using the maximum capacity available for best economical effect
- Multiple TMTplex experiments can be combined by making use of a common scaling sample that is included in all plexes
- Input amounts typically range between 5 µg - 100 µg per condition depending on downstream applications (e.g. enrichment, fractionation)
- TMT10 and TMTpro reagents contain isotope impurities which can be addressed during data analysis and by using an optimized labeling scheme
- Buffers with free amines should be avoided during the labeling reaction as they compete for the labeling reagents leading to incomplete labeling
- Side reactions on Serines, Threonines, Tyrosines and Histidines are possible
Turnaround time
- QC (Pilot) step: 1-2 weeks
- Main TMT multiplexed experiment (3-4 weeks)
Special Workflows using multiplexing
Phosphoproteomics- We only recommend multiplexing for phospho-analyses in combination with
- Fractionation, which leads to significantly deeper analytical depth compared to LFQ
- Specialized enrichment steps such as pTyr antibodies
Thermal Proteome Profiling (login required to access this page)